Help Page of APPred
APPred Webserver
This web server has been designed to assist scientists in predicting and designing antiprotozoal peptides. This page helps the user on how different modules of this web server work. This server's key modules are as follows:
Predict, Design and Protein Scan
Some of the standard terms used in this study are as follows:
Negative Dataset – Peptides with no antiprotozoal activity. Five negative datasets, Non-Antimicrobial peptides, Antiviral peptides, Antibacterial peptides, Antifungal peptides, and Antimicrobial peptides (AMP) excluding antiprotozoal peptides, are used.
Positive Dataset – Peptides with antiprotozoal activity are used.
Feature Selection Method –Feature selection methods i.e SVC-L1 and mRMR are used to select relevant features from a pool of features to build an efficient machine-learning model. The machine learning models built on features selected by mRMR are more robust than the feature selected by SVC-L1.
Prediction Model – Five different classifiers to classify antiprotozoal peptides from the negative class. The prediction models are Decision Tree, Random Forest, Support Vector Machine, Logistic Regression and XGBoost. XgBoost using features selected by mRMR outperformed all other classifiers in classifying antiprotozoal peptides from the negative class.
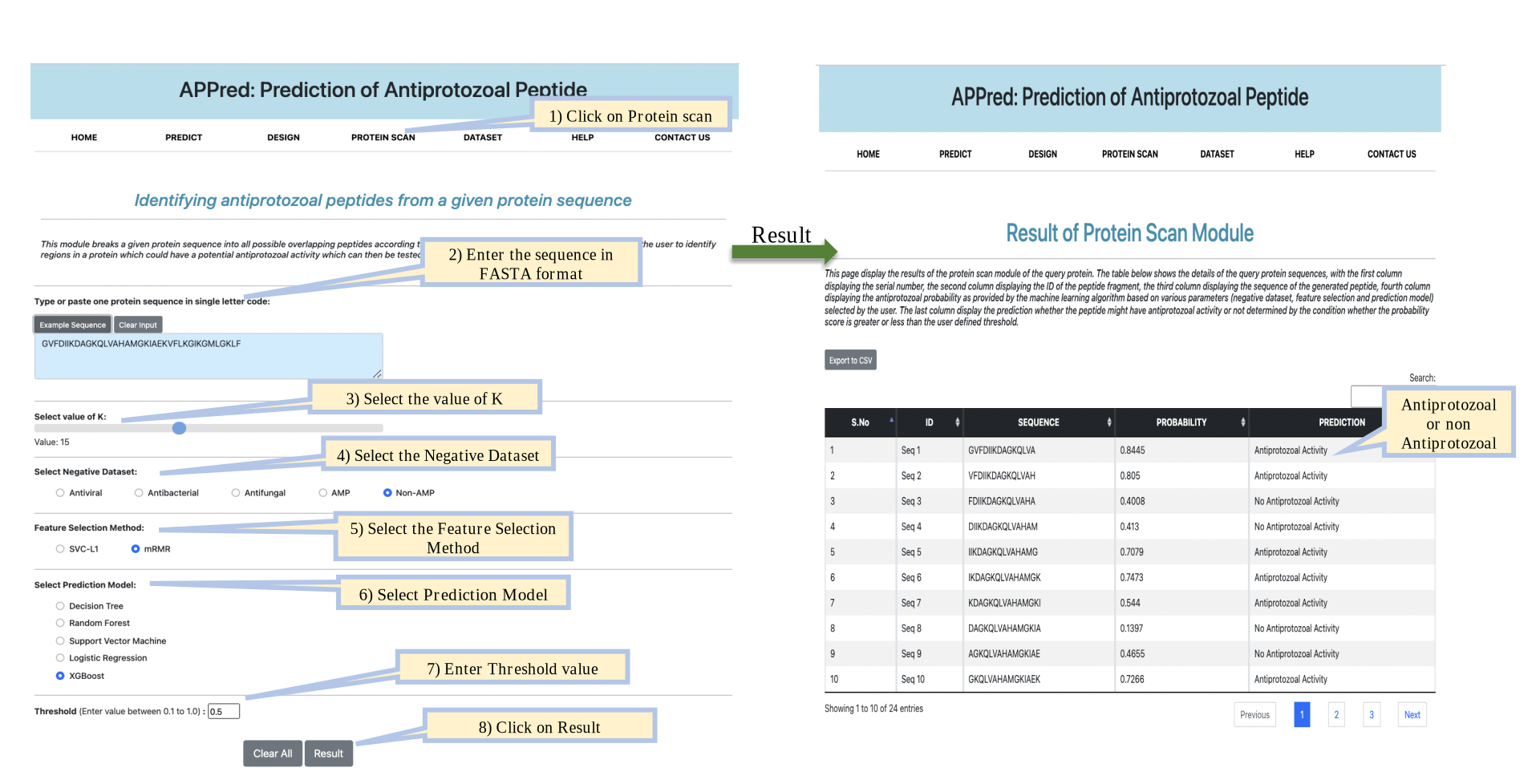
Predict
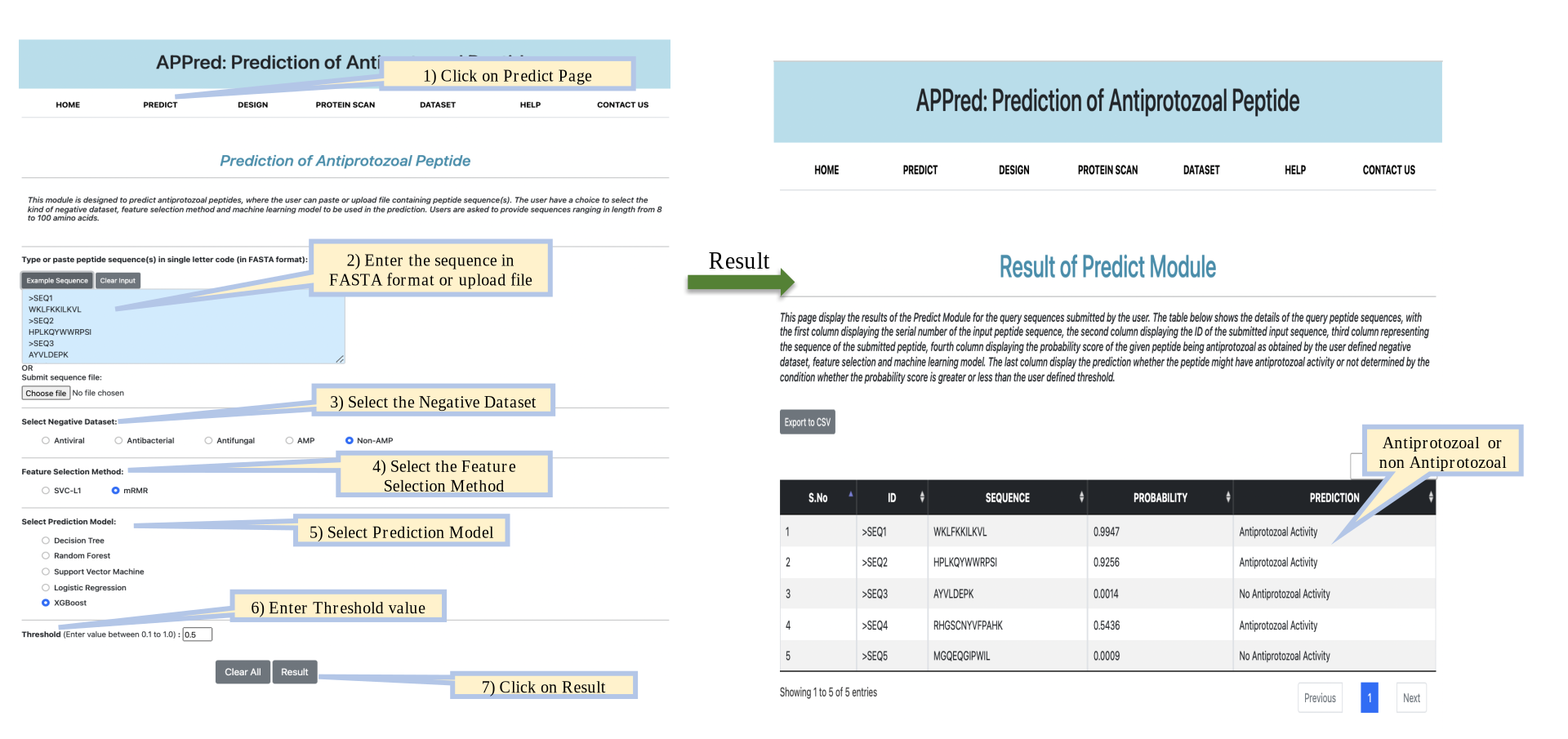
Design
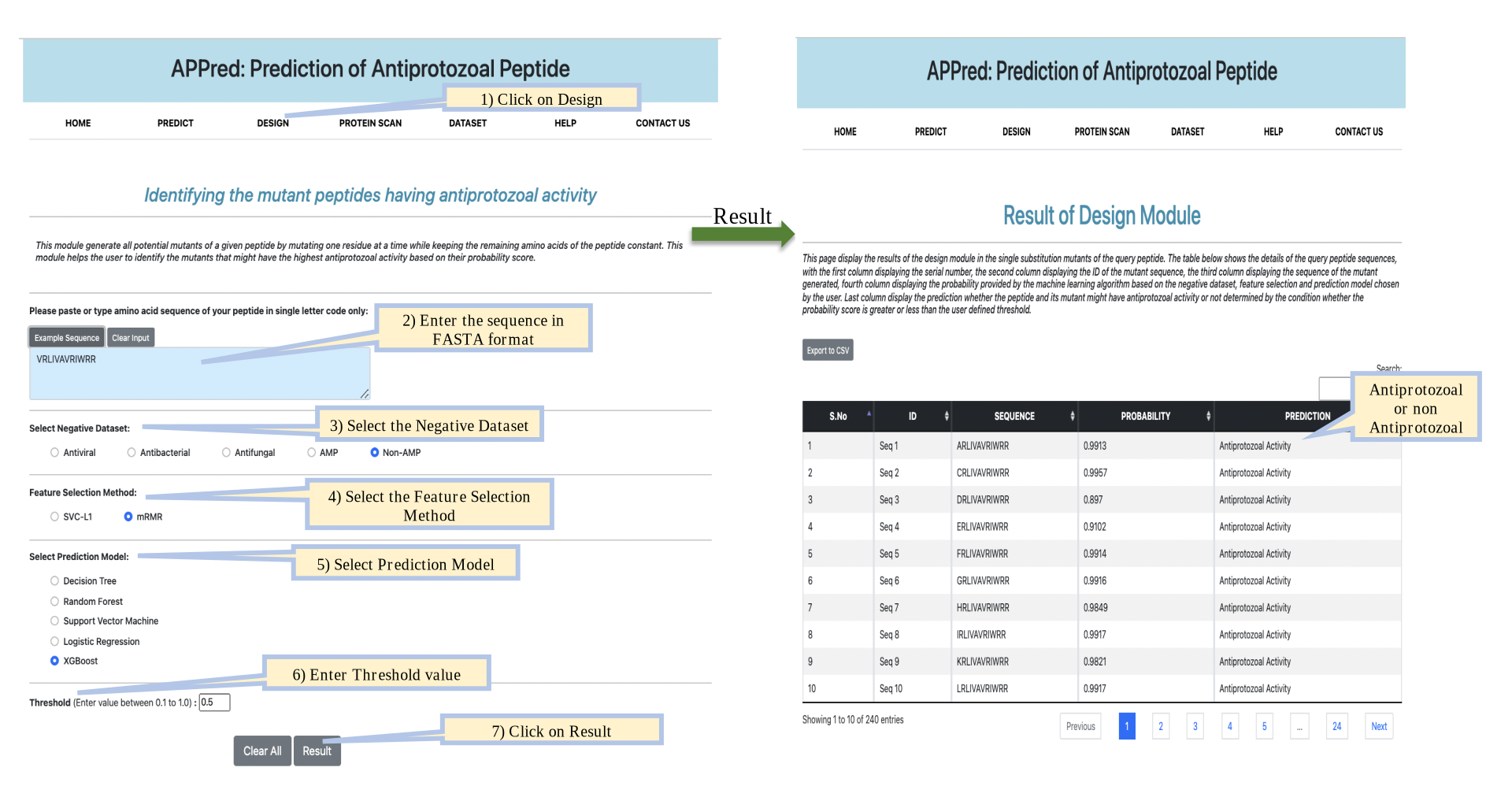
Protein Scan